We develop and deploy cutting-edge molecular techniques to more deeply understand the function of cellular specialization in the nervous system. In particular, we seek clear, actionable explanations for how the cells of the brain go awry in major neuropsychiatric illnesses.
Genomics-based technologies for measuring tissues

We see tremendous opportunity right now to develop new molecular technologies that redefine the scope and scale of what is possible to measure in biology. By combining molecular biology, microfluidics, and organic chemistry, we are trying to build better tools for detailed, information-rich molecular analyses of brain tissues. We recently developed Slide-tags, a method for obtaining high-throughput and spatially resolved single-cell measurements. This technology builds on our Slide-seq platform for spatial transcriptomics and is particularly well suited to application to studying the human brain.
We currently are focused on technologies that can measure synaptic connectivity and plasticity, and methods that detect molecular interactions in situ. We see these new tools as a basis for a new field of tissue biology we call “Tissue Genomics,” which seeks to leverage the comprehensiveness and scale of modern genomics methods to more deeply understand how tissues operate.
Disease Neurobiology
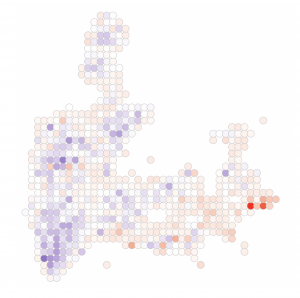
Spatial and single cell genomics technologies provide exciting opportunities to more clearly characterize how tissues respond to disease states. We have three major areas of interest:
New models of neuropsychiatric disease. We have generated mice carrying loss-of-function alleles of four genes recently associated with an elevated risk of developing schizophrenia. The roles of these genes–Xpo7, Cul1, Herc1, and Rb1cc1–are completely unknown in the brain. We are leveraging our genomics tools to characterize their functions and nominate shared pathways for potential therapeutic intervention.
Parkinson’s Disease. We recently developed a strategy for enriching midbrain dopaminergic neuron nuclei from postmortem human tissue, and profiling these nuclei by single-nucleus RNA-seq. We used this approach to molecularly define the population of dopamine neurons most vulnerable to degeneration in Parkinson’s disease. We are now broadening this work to more subjects, and more brain areas, to better understand the neuronal states associated with vulnerability to the disease.
Alzheimer’s Disease. Using rare surgical biopsy samples, we defined cell states in the frontal cortex associated with amyloid accumulation in the human brain. Two particularly intriguing results from this initial work were: 1) oligodendrocytes show gene expression changes indicating an amyloidogenic phenotype and are capable of producing amyloid at levels similar to neurons; and 2) a subset of neurons show a hyperactive phenotype. We are now developing tractable models for studying these novel aspects of Alzheimer’s biology.
Cell type atlases of the mammalian brain
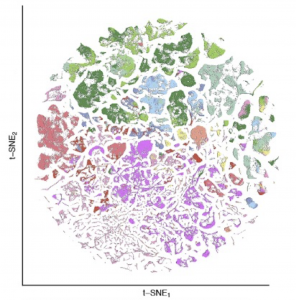
Slide-tags and Slide-seq provide an unprecedented opportunity to define the cytoarchitecture of entire mammalian brains. We recently constructed the first complete atlas of cell types in the mouse brain.
Particularly in midbrain and hindbrain, we identify many specialized neuronal populations, many of which are largely uncharacterized by modern neuroscience.
Recent technological improvements, particularly in Slide-tags, now make it possible to scale our efforts to the (much larger) human brain. We are co-leading a new effort at the Broad Institute–the Center for Human Brain Variation–to characterize the cell types of the human brain, and understand how they vary across individuals.
Novel Computation, software and tools
LIGER
We’ve developed LIGER (Linked Inference of Genomic Experimental Relationships), a new pipeline and R package for comparing and contrasting single-cell datasets across individuals, species, modalities, and other experimental contexts. Learn more about the method and how to use the package here.
Drop-seq
Drop-seq was Evan’s foray into tissue genomics methods development in 2015, and embodies how we think about inventing technologies in the lab today: first, we build an assay that clearly informs if the method is working. Second, using the assay as a guide, we explore ideas widely, aiming to maximize technical simplicity. Third, when we’re successful, we try to disseminate it as widely as possible, to maximize its impact, and get critical feedback on its utility from other scientists.
Here is some (now archival) footage introducing Drop-seq: